DNA Sequencing in the Field – Expanding the Potential of DNA Analysis to Improve Health, Science and National Security
The Problem
Rapid, accurate biological threat identification is critical for making informed decisions that protect our nation. DNA sequencing – the process of “reading” a sample’s genetic blueprint – is the integral first step in organism identification which has widespread applications in disease prevention and cure development, advancement of agriculture and life sciences and defense against biological weapons. Although DNA sequencing has become a well-established industry practice, typical methods of sample collection and transportation to a dedicated lab facility for analysis consumes valuable time when public health and safety are at stake. Further increasing turnaround time, sequencing and analysis often requires a resource-intensive high performance computing (HPC) infrastructure to generate actionable insight.
Our Approach
In mission-critical applications such as biosurveillance—the tracking and detection of potentially harmful biological agents for national defense and public health missions—time is everything. Noblis has developed an end-to-end field-portable genetic sequencing and analysis system to decrease response times.
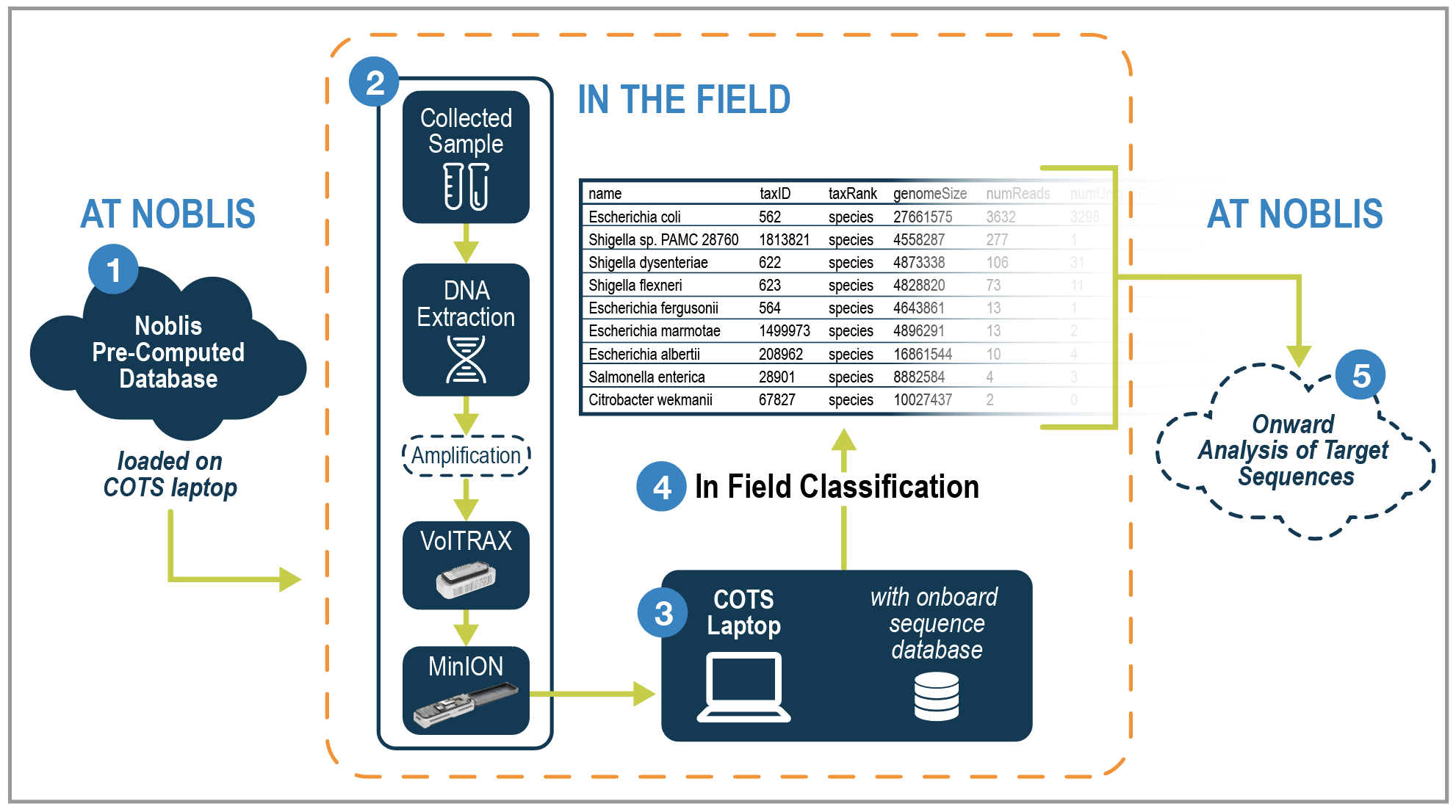
The Noblis research team’s solution (depicted in Figure 1):
- Enables rapid metagenomic classification analysis in the field without an internet connection or in line power
- Provides a complete end-to-end sample to answer analysis in less than 3.5 hours (versus facilities-based sample analysis in about 24 hours)
- Is equipped with field portable miniaturized sequencing technology to expand the usability and efficiency of field based genetic sequencing
- Is preloaded with databases created by Noblis subject matter experts representing large genomic datasets that have been reduced into a footprint small enough to fit on a commercial-off-the-shelf (COTS) laptop
Figure 1 – Noblis Field-Portable DNA Sequencing Solution
Our process includes 1) loading pre-computed databases onto a COTS laptop, 2) in field sample collection and processing including preparation, DNA extraction, purification, sequencing, basecalling, 3) & 4) in field classification without internet or inline power leveraging pre-installed databases, and 5) optional additional anslysis of target sequences back at Noblis facilities.
Watch Our Video to See the Solution in Action
Accomplishments
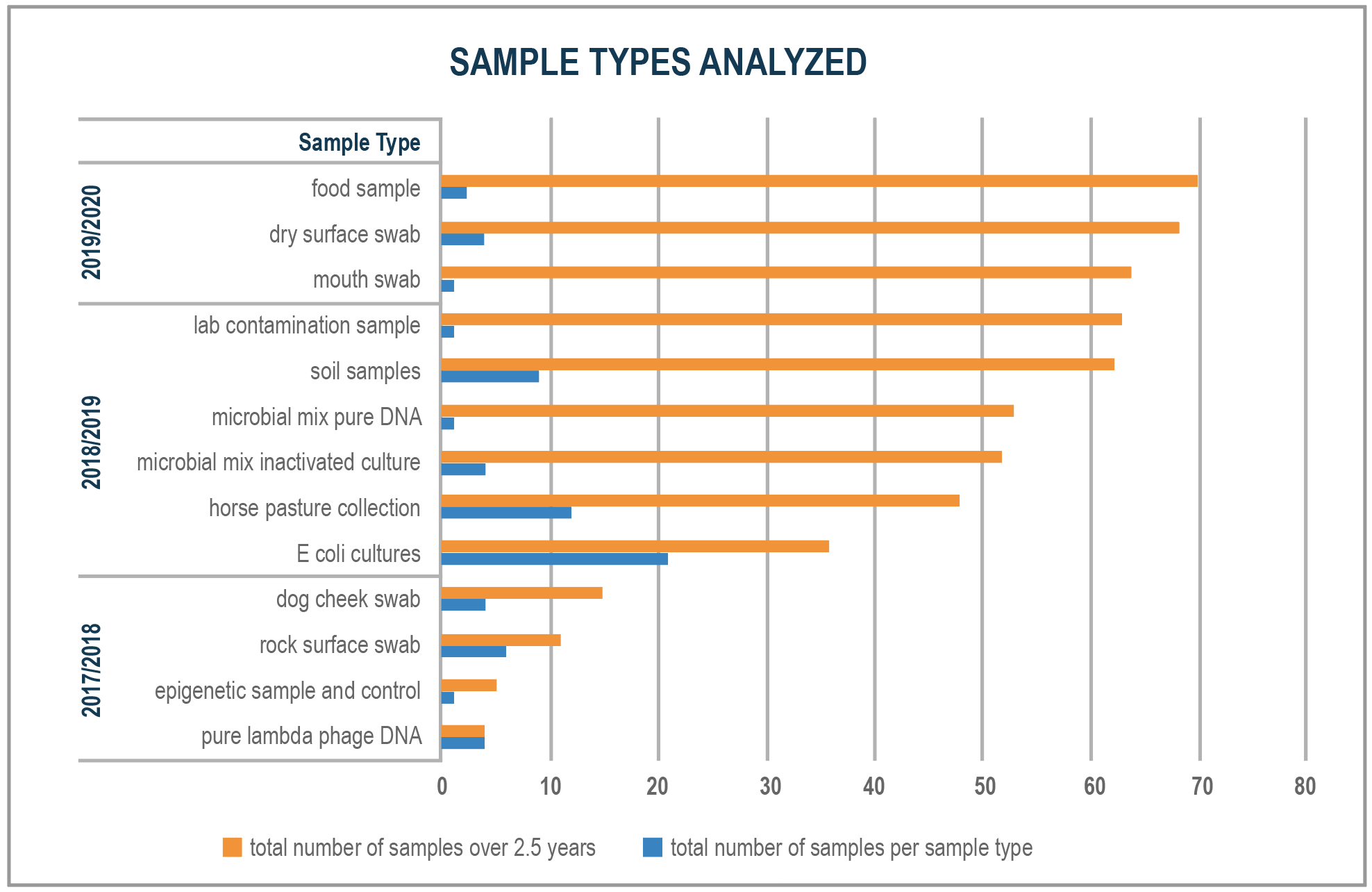
Noblis is currently completing sequencing in about 3.5 hours and is working to decrease that time further.Industry standard sample collection and preparation—including transportation to a lab facility for processing on conventional equipment, a step not practicable in many real-world applications—has a minimum 24-hour turnaround time. Since developing this portable system in 2017, Noblis has successfully performed whole molecule sequencing runs on about 70 samples (Figure 2) to prove sensitivity and specificity. Metagenomic analysis has been performed on environmental samples including:
- pond water samples
- soil samples
- dry surface swabs
- dog cheek swabs
- pure cultures
In addition, epigenetic analysis has been performed on pure DNA samples.
Figure 2 – Sample Types Analyzed
Inquiries
Noblis Sponsored Research projects are conceptualized and developed with the public interest and government requirements at the forefront of our envisioned outcomes. We don’t believe in a “one size fits all” approach and can work with your agency to determine how to best tailor our solutions and expertise to your unique needs.
- To learn more about this project and how it could contribute to your mission, contact us here. View our solution spec sheet here.
- To learn more about the Noblis Sponsored Research program, visit noblis.org/r-d.
- To learn more about Noblis capabilities and solutions in life sciences and bioinformatics, visit bioportal.noblis.org.